The iml package can now handle bigger datasets. Earlier
problems with exploding memory have been fixed for
FeatureEffect, FeatureImp and
Interaction. It’s also possible now to compute
FeatureImp and Interaction in parallel. This
document describes how.
First we load some data, fit a random forest and create a Predictor object.
set.seed(42)
library("iml")
library("randomForest")
#> randomForest 4.7-1.2
#> Type rfNews() to see new features/changes/bug fixes.
data("Boston", package = "MASS")
rf <- randomForest(medv ~ ., data = Boston, n.trees = 10)
X <- Boston[which(names(Boston) != "medv")]
predictor <- Predictor$new(rf, data = X, y = Boston$medv)Going parallel
Parallelization is supported via the {future} package. All you need
to do is to choose a parallel backend via
future::plan().
library("future")
library("future.callr")
# Creates a PSOCK cluster with 2 cores
plan("callr", workers = 2)Now we can easily compute feature importance in parallel. This means that the computation per feature is distributed among the 2 cores I specified earlier.
imp <- FeatureImp$new(predictor, loss = "mae")
library("ggplot2")
#>
#> Attaching package: 'ggplot2'
#> The following object is masked from 'package:randomForest':
#>
#> margin
plot(imp)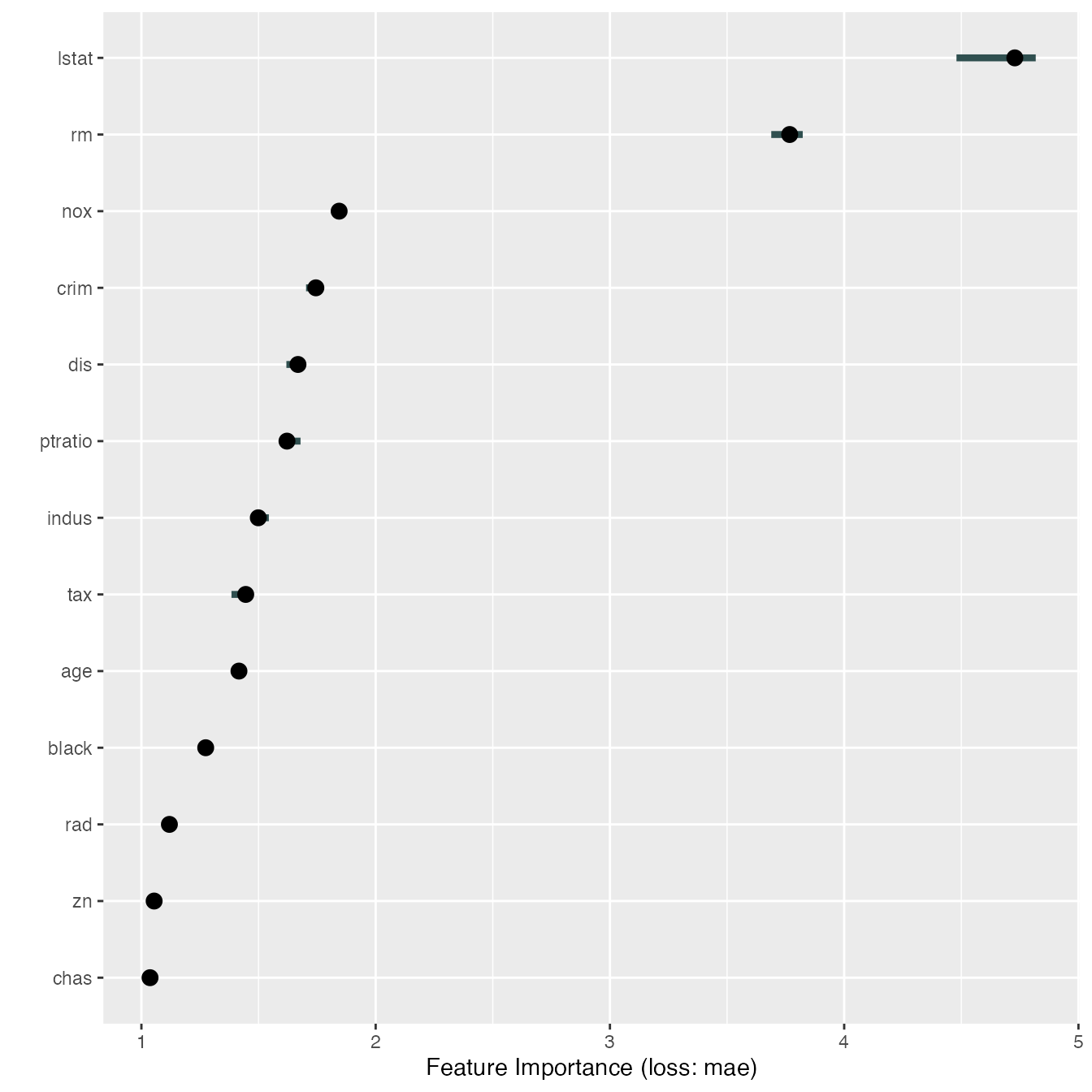
That wasn’t very impressive, let’s actually see how much speed up we get by parallelization.
bench::system_time({
plan(sequential)
FeatureImp$new(predictor, loss = "mae")
})
#> process real
#> 2.09s 2.06s
bench::system_time({
plan("callr", workers = 2)
FeatureImp$new(predictor, loss = "mae")
})
#> process real
#> 145.53ms 2.05sA little bit of improvement, but not too impressive. Parallelization is more useful in the case where the model uses a lot of features or where the feature importance computation is repeated more often to get more stable results.
bench::system_time({
plan(sequential)
FeatureImp$new(predictor, loss = "mae", n.repetitions = 10)
})
#> process real
#> 3.49s 3.44s
bench::system_time({
plan("callr", workers = 2)
FeatureImp$new(predictor, loss = "mae", n.repetitions = 10)
})
#> process real
#> 160.4ms 2.99sInteraction
Here the parallel computation is twice as fast as the sequential computation of the feature importance.
The parallelization also speeds up the computation of the interaction statistics:
bench::system_time({
plan(sequential)
Interaction$new(predictor, grid.size = 15)
})
#> process real
#> 7.61s 7.49s
bench::system_time({
plan("callr", workers = 2)
Interaction$new(predictor, grid.size = 15)
})
#> process real
#> 157.13ms 5.16sFeature Effects
Same for FeatureEffects:
bench::system_time({
plan(sequential)
FeatureEffects$new(predictor)
})
#> process real
#> 1.09s 1.07s
bench::system_time({
plan("callr", workers = 2)
FeatureEffects$new(predictor)
})
#> process real
#> 694.86ms 2.95s