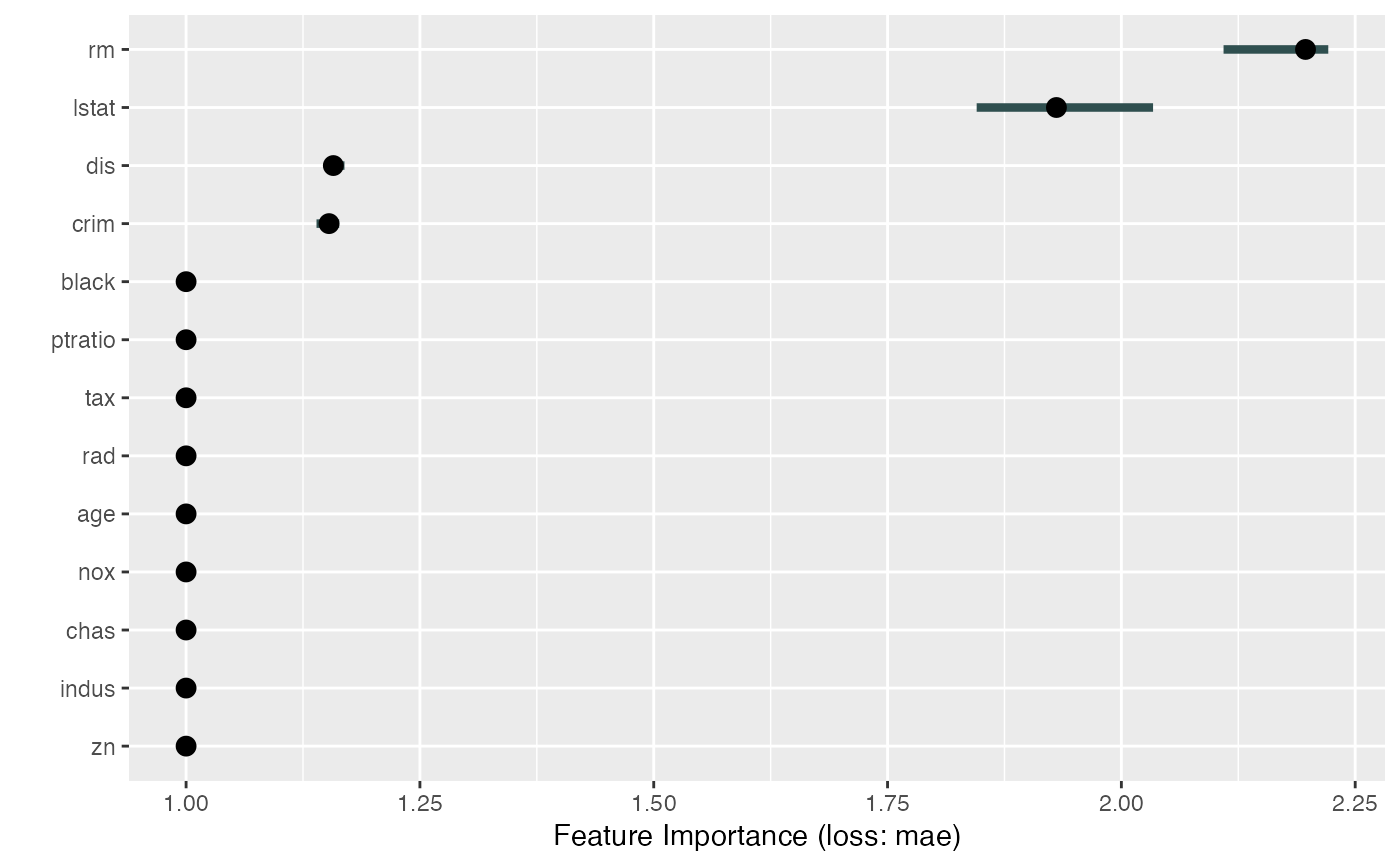
plot.FeatureImp() plots the feature importance results of a FeatureImp
object.
# S3 method for class 'FeatureImp'
plot(x, sort = TRUE, ...)Arguments
- x
A FeatureImp object
- sort
logical. Should the features be sorted in descending order? Defaults to TRUE.
- ...
Further arguments for the objects plot function
Value
ggplot2 plot object
Details
The plot shows the importance per feature.
When n.repetitions in FeatureImp$new was larger than 1, then we get
multiple importance estimates per feature. The importance are aggregated and
the plot shows the median importance per feature (as dots) and also the
90%-quantile, which helps to understand how much variance the computation has
per feature.
See also
Examples
library("rpart")
# We train a tree on the Boston dataset:
data("Boston", package = "MASS")
tree <- rpart(medv ~ ., data = Boston)
y <- Boston$medv
X <- Boston[-which(names(Boston) == "medv")]
mod <- Predictor$new(tree, data = X, y = y)
# Compute feature importances as the performance drop in mean absolute error
imp <- FeatureImp$new(mod, loss = "mae")
# Plot the results directly
plot(imp)